MetaCyto: a tool for meta-analysis of flow cytometry and mass cytometry data
Meta-analysis of existing data across different studies offers multiple benefits, including increased statistical power and more robust conclusions. These benefits have been shown by many studies in areas such as genomics, cancer biology, and clinical research, and have led to important new biomedical findings.
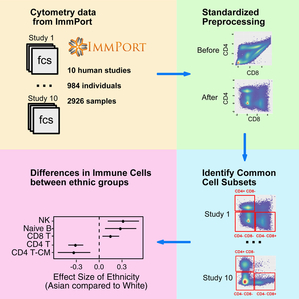
With the recent advances in high-throughput cytometry technologies, the immune system can be characterized at the single-cell level with up to 45 parameters, minimizing the technical limitations and allowing capture of more valuable information from immunology studies. Open science initiatives have led to more of this type of research data being accessible, and the availability of shared cytometry data, including data from flow cytometry and mass cytometry (CyTOF), is growing exponentially. Notably, the ImmPort database, a repository for immunology-related research and clinical trials, provides numerous studies with thousands of cytometry datasets.
However, meta-analysis of cytometry datasets remained particularly challenging. Different studies use diverse sets of protein markers and fluorophore/isotope combinations. The detected values of the same marker are inconsistent between studies because of different cytometer configurations or operators. In addition, the high dimensionality of cytometry data, especially CyTOF data, makes manual gating-based meta-analysis difficult and time-consuming.
A research team headed by Atul Butte at the University of California, San Francisco has developed a new computational pipeline named MetaCyto to enable automated meta-analysis of cytometry datasets. MetaCyto allows researchers to analyze thousands of heterogeneous flow cytometry or CyTOF data within minutes. Such analysis would take researchers months if done manually.
“This is the first time a large-scale meta-analysis of cytometry data has been done. It shows that heterogeneous cytometry data from independent studies can be combined to provide new insights,” says Zicheng Hu, the first author of the study.
Using MetaCyto, the team performed a joint analysis of 2,926 peripheral blood mononuclear cell (PBMC) samples from ten human immunology cytometry datasets. The meta-analysis approach allows the team to discover new differences in immune cells between demographic groups. The tool is available in _ImmPort Galaxy _ (https://immportgalaxy.org/) and as an R package (https://bioconductor.org/packages/release/bioc/html/MetaCyto.html) and as a galaxy tool (https://immportgalaxy.org/).
Funding for MetaCyto provided by the National Institutes of Health (NIH), National Institutes of Allergy and Infectious Disease, under the ImmPort contract HHSN316201200036W